import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import dash_bio
import scipy
import mathdata = pd.read_parquet('max_gve.parquet.gzip')
data| chrom | pos | name | abs_max_effect | max_column | ave_effect | |
|---|---|---|---|---|---|---|
| 0 | 1 | 174946171 | rs147831690 | 0.027101 | nrnlsc_H3K79me1 | 0.000700 |
| 1 | 1 | 174946180 | rs1389168246 | 0.024842 | nrnlsc_H3K79me1 | 0.000408 |
| 2 | 1 | 174946196 | rs1374951931 | 0.072019 | nrnlsc_H3K79me1 | 0.001219 |
| 3 | 1 | 174946202 | rs948828419 | 0.033973 | nrnlsc_H3K79me1 | 0.001874 |
| 4 | 1 | 174946211 | rs896018724 | 0.064587 | nrnlsc_H3K79me1 | 0.001534 |
| ... | ... | ... | ... | ... | ... | ... |
| 547681 | 2 | 156681080 | rs747765225 | 0.009990 | SKNSH_ARNTL | 0.000462 |
| 547682 | 2 | 156681082 | rs994964368 | 0.009063 | cdtn_H3K4me3 | 0.000822 |
| 547683 | 2 | 156681084 | rs1028224262 | 0.008285 | SKNSH_ARNTL | 0.000428 |
| 547684 | 2 | 156681085 | rs541069773 | 0.016144 | SKNSH_ARNTL | 0.000941 |
| 547685 | 2 | 156681113 | rs1432010733 | 0.016751 | MOC_NP_ATAC | 0.000612 |
16476152 rows × 6 columns
dfs = []
for i in range(10):
print(i)
df = data.iloc[:1000, :]
dfs.append(df)
print(df.head())0
1
2
3
4
5
6
7
8
9
chrom pos name abs_max_effect max_column ave_effect
0 1 174946171 rs147831690 0.027101 nrnlsc_H3K79me1 0.000700
1 1 174946180 rs1389168246 0.024842 nrnlsc_H3K79me1 0.000408
2 1 174946196 rs1374951931 0.072019 nrnlsc_H3K79me1 0.001219
3 1 174946202 rs948828419 0.033973 nrnlsc_H3K79me1 0.001874
4 1 174946211 rs896018724 0.064587 nrnlsc_H3K79me1 0.001534dfs = pd.concat(dfs)dfs| chrom | pos | name | abs_max_effect | max_column | ave_effect | |
|---|---|---|---|---|---|---|
| 0 | 1 | 174946171 | rs147831690 | 0.027101 | nrnlsc_H3K79me1 | 0.000700 |
| 1 | 1 | 174946180 | rs1389168246 | 0.024842 | nrnlsc_H3K79me1 | 0.000408 |
| 2 | 1 | 174946196 | rs1374951931 | 0.072019 | nrnlsc_H3K79me1 | 0.001219 |
| 3 | 1 | 174946202 | rs948828419 | 0.033973 | nrnlsc_H3K79me1 | 0.001874 |
| 4 | 1 | 174946211 | rs896018724 | 0.064587 | nrnlsc_H3K79me1 | 0.001534 |
| ... | ... | ... | ... | ... | ... | ... |
| 995 | 1 | 174949547 | rs1573971060 | 0.042717 | MOC_NP_ATAC | 0.001534 |
| 996 | 1 | 174949551 | rs1158195944 | 0.005916 | SKNSH_H3F3A | 0.000336 |
| 997 | 1 | 174949556 | rs191506112 | 0.006997 | ACC_NP_ATAC | 0.000302 |
| 998 | 1 | 174949557 | rs557939733 | 0.005723 | ACC_NP_ATAC | 0.000226 |
| 999 | 1 | 174949559 | rs551299464 | 0.007402 | MiddleFrGy_DNase | 0.000275 |
10000 rows × 6 columns
data = pd.read_csv('max_gve.csv')
data['chrom'][data.chrom=='X'] = 23
data['chrom'][data.chrom=='Y'] = 24
data['chrom'][data.chrom=='16.0'] = 16
data['chrom'] = data.chrom.astype(int)
data| chrom | pos | name | max_effect | max_column | ave_effect | |
|---|---|---|---|---|---|---|
| 0 | 1 | 238807165 | rs1207962803 | 0.015620 | brnmec_DNase | 0.000920 |
| 1 | 1 | 238807165 | rs1242649293 | 0.081236 | nrnlsc_H3K9ac | -0.002660 |
| 2 | 1 | 238807167 | rs997302978 | 0.002643 | ACC_NP_ATAC | -0.001335 |
| 3 | 1 | 238807168 | rs1270214146 | 0.045428 | nrnlsc_H3K79me1 | -0.000773 |
| 4 | 1 | 238807168 | rs1270214146 | 0.083614 | nrnlsc_H3K79me1 | 0.001402 |
| ... | ... | ... | ... | ... | ... | ... |
| 16652518 | 2 | 156681080 | rs747765225 | 0.006044 | PMC_NN_ATAC | -0.000144 |
| 16652519 | 2 | 156681082 | rs994964368 | 0.000293 | MeT_NN_ATAC | -0.000791 |
| 16652520 | 2 | 156681084 | rs1028224262 | 0.004548 | VPC_NN_ATAC | -0.000109 |
| 16652521 | 2 | 156681085 | rs541069773 | 0.011259 | VPC_NN_ATAC | -0.000122 |
| 16652522 | 2 | 156681113 | rs1432010733 | 0.010121 | nrnlsc_H3K4me1 | 0.000242 |
16652523 rows × 6 columns
data.chrom.unique()array([ 1, 22, 3, 12, 16, 17, 13, 10, 11, 2, 19, 14, 15, 21])neg = -(data.max_effect.values)
exp = np.exp(neg)
data['exp'] = exp
zscores = scipy.stats.zscore(data.ave_effect.values)
data['zscore'] = zscores
pvalues = scipy.stats.norm.sf(zscores)
data['p'] = pvalues
data| chrom | pos | name | max_effect | max_column | ave_effect | exp | zscore | p | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 238807165 | rs1207962803 | 0.015620 | brnmec_DNase | 0.000920 | 0.984502 | 0.310131 | 0.378231 |
| 1 | 1 | 238807165 | rs1242649293 | 0.081236 | nrnlsc_H3K9ac | -0.002660 | 0.921976 | -0.931275 | 0.824144 |
| 2 | 1 | 238807167 | rs997302978 | 0.002643 | ACC_NP_ATAC | -0.001335 | 0.997360 | -0.471795 | 0.681464 |
| 3 | 1 | 238807168 | rs1270214146 | 0.045428 | nrnlsc_H3K79me1 | -0.000773 | 0.955588 | -0.276915 | 0.609077 |
| 4 | 1 | 238807168 | rs1270214146 | 0.083614 | nrnlsc_H3K79me1 | 0.001402 | 0.919786 | 0.477353 | 0.316555 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 16652518 | 2 | 156681080 | rs747765225 | 0.006044 | PMC_NN_ATAC | -0.000144 | 0.993974 | -0.058674 | 0.523394 |
| 16652519 | 2 | 156681082 | rs994964368 | 0.000293 | MeT_NN_ATAC | -0.000791 | 0.999707 | -0.283229 | 0.611499 |
| 16652520 | 2 | 156681084 | rs1028224262 | 0.004548 | VPC_NN_ATAC | -0.000109 | 0.995462 | -0.046438 | 0.518520 |
| 16652521 | 2 | 156681085 | rs541069773 | 0.011259 | VPC_NN_ATAC | -0.000122 | 0.988804 | -0.050923 | 0.520306 |
| 16652522 | 2 | 156681113 | rs1432010733 | 0.010121 | nrnlsc_H3K4me1 | 0.000242 | 0.989930 | 0.075191 | 0.470031 |
16652523 rows × 9 columns
data.to_csv('abs_max_ave_effect_pvalue.csv', index=False)data = pd.read_csv('tbln_H3K27ac_kmeans0.csv')
data['chrom'][data.chrom=='X'] = 23
data['chrom'][data.chrom=='Y'] = 24
data['chrom'][data.chrom=='16.0'] = 16
data['chrom'] = data.chrom.astype(int)
data| chrom | pos | name | tbln_H3K27ac | diff | abs_diff | |
|---|---|---|---|---|---|---|
| 0 | 10 | 84139514.0 | rs932821133 | 0.105500 | 0.105500 | 0.105500 |
| 1 | 10 | 84139543.0 | rs1229596357 | -0.183456 | -0.183456 | 0.183456 |
| 2 | 10 | 84139573.0 | rs1481839079 | 0.143182 | 0.143182 | 0.143182 |
| 3 | 10 | 84140064.0 | rs1452183496 | 0.136644 | 0.136644 | 0.136644 |
| 4 | 10 | 84140294.0 | rs1429667538 | 0.100358 | 0.100358 | 0.100358 |
| ... | ... | ... | ... | ... | ... | ... |
| 726226 | 3 | 194089744.0 | rs1351149363 | -0.218239 | -0.218239 | 0.218239 |
| 726227 | 3 | 194089846.0 | rs890061713 | -0.116548 | -0.116548 | 0.116548 |
| 726228 | 3 | 194089911.0 | rs1246306491 | -0.164336 | -0.164336 | 0.164336 |
| 726229 | 3 | 194090129.0 | rs1181593567 | -0.111510 | -0.111510 | 0.111510 |
| 726230 | 3 | 194095916.0 | rs1009672135 | -0.116104 | -0.116104 | 0.116104 |
726231 rows × 6 columns
neg = -(data.tbln_H3K27ac.values)
exp = np.exp(neg)
data['exp'] = exp
zscores = scipy.stats.zscore(data.tbln_H3K27ac.values)
data['zscore'] = zscores
pvalues = scipy.stats.norm.sf(zscores)
data['p'] = pvalues
data| chrom | pos | name | tbln_H3K27ac | diff | abs_diff | exp | |
|---|---|---|---|---|---|---|---|
| 0 | 10 | 84139514.0 | rs932821133 | 0.105500 | 0.105500 | 0.105500 | 0.899874 |
| 1 | 10 | 84139543.0 | rs1229596357 | -0.183456 | -0.183456 | 0.183456 | 1.201363 |
| 2 | 10 | 84139573.0 | rs1481839079 | 0.143182 | 0.143182 | 0.143182 | 0.866597 |
| 3 | 10 | 84140064.0 | rs1452183496 | 0.136644 | 0.136644 | 0.136644 | 0.872280 |
| 4 | 10 | 84140294.0 | rs1429667538 | 0.100358 | 0.100358 | 0.100358 | 0.904514 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 726226 | 3 | 194089744.0 | rs1351149363 | -0.218239 | -0.218239 | 0.218239 | 1.243884 |
| 726227 | 3 | 194089846.0 | rs890061713 | -0.116548 | -0.116548 | 0.116548 | 1.123611 |
| 726228 | 3 | 194089911.0 | rs1246306491 | -0.164336 | -0.164336 | 0.164336 | 1.178611 |
| 726229 | 3 | 194090129.0 | rs1181593567 | -0.111510 | -0.111510 | 0.111510 | 1.117965 |
| 726230 | 3 | 194095916.0 | rs1009672135 | -0.116104 | -0.116104 | 0.116104 | 1.123113 |
726231 rows × 7 columns
plt.hist(data.tbln_H3K27ac)(array([2.40000e+01, 5.00000e+02, 1.43580e+04, 3.43638e+05, 0.00000e+00,
3.38365e+05, 2.81640e+04, 1.12700e+03, 4.00000e+01, 1.50000e+01]),
array([-0.74844134, -0.58586169, -0.42328205, -0.2607024 , -0.09812275,
0.0644569 , 0.22703654, 0.38961619, 0.55219584, 0.71477548,
0.87735513]),
<BarContainer object of 10 artists>)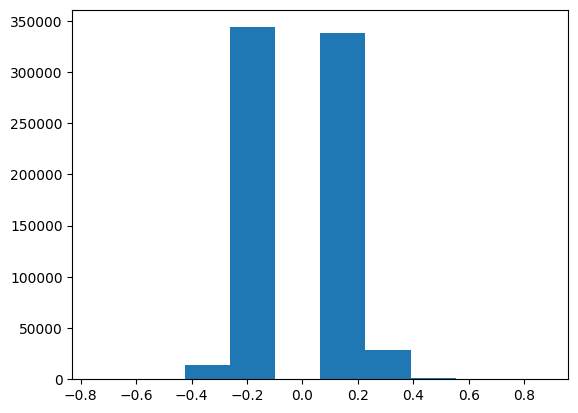
# scipy.stats.t.sf(data.ITC_NN_ATAC.values, df=2)# scipy.stats.t.sf(data.ITC_NN_ATAC.values, df=3)| chrom | pos | name | tbln_H3K27ac | diff | abs_diff | exp | zscore | p | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 84139514.0 | rs932821133 | 0.105500 | 0.105500 | 0.105500 | 0.899874 | 0.658540 | 0.255096 |
| 1 | 10 | 84139543.0 | rs1229596357 | -0.183456 | -0.183456 | 0.183456 | 1.201363 | -1.181742 | 0.881346 |
| 2 | 10 | 84139573.0 | rs1481839079 | 0.143182 | 0.143182 | 0.143182 | 0.866597 | 0.898519 | 0.184454 |
| 3 | 10 | 84140064.0 | rs1452183496 | 0.136644 | 0.136644 | 0.136644 | 0.872280 | 0.856886 | 0.195754 |
| 4 | 10 | 84140294.0 | rs1429667538 | 0.100358 | 0.100358 | 0.100358 | 0.904514 | 0.625787 | 0.265727 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 726226 | 3 | 194089744.0 | rs1351149363 | -0.218239 | -0.218239 | 0.218239 | 1.243884 | -1.403260 | 0.919730 |
| 726227 | 3 | 194089846.0 | rs890061713 | -0.116548 | -0.116548 | 0.116548 | 1.123611 | -0.755621 | 0.775062 |
| 726228 | 3 | 194089911.0 | rs1246306491 | -0.164336 | -0.164336 | 0.164336 | 1.178611 | -1.059971 | 0.855421 |
| 726229 | 3 | 194090129.0 | rs1181593567 | -0.111510 | -0.111510 | 0.111510 | 1.117965 | -0.723538 | 0.765325 |
| 726230 | 3 | 194095916.0 | rs1009672135 | -0.116104 | -0.116104 | 0.116104 | 1.123113 | -0.752797 | 0.774214 |
726231 rows × 9 columns
data.chrom.unique()array([10, 1, 16, 23, 15, 21, 2, 6, 7, 5, 12, 19, 8, 3, 9, 4, 11,
24, 13, 14, 17, 22, 20, 18])data.to_csv('data_with_pvalue.csv', index=False)dash_bio.ManhattanPlot(dataframe=df, chrm='CHR', bp='BP', snp='SNP', p='P')